|
“We’re all connected
somehow… somewhere… some point in time.”
mtDNA Genetics
I. Maternal
Lineage – U2B

mtDNA Haplogroup U Origins
Haplogroup
U is believed to have arisen somewhere in Europe or the Near
East approximately 55,000 years before present. Haplogroup U
is a group of individuals who descend from a woman in the R branch of the
tree. Because of the great genetic diversity found in haplogroup U, it is
likely that she lived around 50,000-55,000 years ago. Her descendants gave rise to several
different subgroups, some of which exhibit very specific geographic
homelands. The very old age of these subgroups has led to a wide
distribution; today they harbor specific European, northern African, and
Indian components, and are found in Arabia, the northern Caucasus
Mountains, and throughout the Near East. Haplogroup U is found throughout Europe, and contains many subgroups, each reflecting
unique geography and history. Among its subgroups is Haplogroup K.
While
some members of haplogroup U headed north into Scandinavia, or south into
North Africa, most members of haplogroup U stem from a group that moved
northward out of the Near East. These
women crossed the rugged Caucasus Mountains in Southern Russia, and moved
on to the steppes of the Black Sea. These
individuals represent movements from the Black Sea steppes west into
regions that comprise the present-day Baltic States and western Eurasia. This grassland then served as the home base
for subsequent movements north and west. Today, members of these lineages
are found in Europe and the eastern Mediterranean
at frequencies of almost seven percent of the population.
Haplogroup U is subdivided into Haplogroups
U1-U8 and Haplogroup K.
Haplogroup U1
Haplogroup
U1 is found in about 14.4 percent of Armenians/Georgians in the Caucasus region, and about 2.3 percent of Indians,
1.2 percent of Estonians, Russians, and Slovaks.
Haplogroup U2
The
distribution of the three sister clades within haplogroup U2 (U2a, U2b, and U2c) is essentially restricted to the Indo-Pakistani regions.
They have not been observed in Europe and the Near
East and, according to our data, they are absent in the
Iranian plateau and Central Asian populations. They are, however, common in
populations from Pakistan
and India.
The estimated coalescence times for these haplogroups are: 45,700 ± 14,400
years for U2a, 35,900 ±
9,000 years for U2b, and 45,200 ± 10,400
years for U2c.
Haplogroup U3
U3
has also been found with a comparatively higher frequency in Northwest
Africa and might have followed the same route as U6, however, as its
star-like expansion in the Caucasus has been dated around 30,000 yr BP, it
most probably reached Africa in a posterior expansion. This out of Africa and back again hypothesis has also been
suggested for Y-chromosome lineages. Subsequent Neolithic and historic
expansions have doubtlessly reshaped the human genetic pool in wide
geographic areas but mainly as limited gene flow, not admixture, between
populations. Consequently, the continental origin of the major haplogroups
can still be detected and the earliest human routes inferred through them.
Haplogroup U4
Haplogroup
U4 has its origin in the Upper Palaeolithic,
dating to approximately 25,000 years ago. It is widely distributed in
Europe, and has been implicated in the expansion of modern humans into Europe occurring before the Last Glacial Maximum.
Haplogroup U5
Haplogroup
U5 is restricted to Finland
in its variation. This is likely the result of the significant
geographical, linguistic, and cultural isolation of the Finnish
populations, which would have restricted geographic distribution of this
subgroup and kept it fairly isolated genetically. The Saami, reindeer
hunters who follow the herds from Siberia to Scandinavia each season, also
have U5 lineages in their population, indicating that these may have been
introduced during their movements into these northern territories. The U5 lineage is
found as far south as the Near East,
though at much smaller frequencies and at much lower diversity. Because
these individuals contain lineages that first evolved in Europe, their
presence in the Near East is the result of a back-migration of people who
left northern Europe and headed south, as
though retracing the migratory paths of their own ancestors.
Haplogroup U6
Another
interesting subgroup is U6, which branched off from haplogroup R while
still in the Middle East. Haplogroup U6,
is common in North Africa, and may suggest a "reverse migration"
from Europe. Today U6 individuals are
found in around ten percent of people living in North
Africa. Despite the great geographic distances between
subgroups U5 and U6, all members still share the same maternal ancestral
line that gave rise to the haplogroup U ancestral clan.
Haplogroup U7
Many
European populations lack Haplogroup U7, but its frequency climbs over 4%
in the Near East and up to 5% in Pakistan, reaching nearly 10%
level in Iranians. In India,
haplogroup U7 frequency peaks at over 12% in Gujarat, the westernmost state
of India, while for the
whole of India
its frequency stays around 2%. Expansion times and haplotype diversities
for the Indian and Near and Middle Eastern U7 mtDNAs are strikingly
similar. The possible homeland of this haplogroup spans Indian Gujarat and Iran
because from there its frequency declines steeply both to the east and to
the west. If the origin were in Iran
rather than in India,
then its equally high frequency as well as diversity in Gujarat favors a
scenario whereby U7 has been introduced to the coastal western India
either very early, or by multiple founders.
Haplogroup U8
The
Basques have the most ancestral phylogeny in Europe for the mitochondrial
haplogroup U8a, a rare subgroup of U8, placing the Basque origin of this
lineage in the Upper Palaeolithic. The
lack of U8a lineages in Africa suggests that their ancestors may have
originated from West Asia.
Haplogroup K
Haplogroup
K is a human mitochondrial DNA (mtDNA) haplogroup. Haplogroup K is part of
the larger haplogroup
U. It is a mostly
Eurasian haplotype, and is believed to have first appeared when human
populations expanded through Europe after
the last glacial maximum in 16,000 BC.
Approximately
32% of the haplotypes of modern people with Ashkenazi Jewish ancestry are
in haplogroup K.
In
his popular book The Seven Daughters of Eve, Bryan Sykes named the
originator of this mtDNA haplogroup Katrine.
This
haplogroup is the final destination of a genetic journey that began some
150,000 years ago with an ancient mtDNA haplogroup called L3. Haplogroup L3
occurs only in Africa, but on that
continent its derivatives are found nearly everywhere. L3's subclades are
most prevalent in East Africa. This
ancient lineage reflects an early divergence from humanity's common genetic
coalescence point. "Mitochondrial Eve," the common ancestor of
all living humans, was born in Africa some
150,000 years ago. All existing mtDNA diversity began with Eve and it
remains greatest, and subsequently oldest, in Africa.
Y chromosome polymorphisms on the male line of descent also point to an
African origin for all humans , but our male common ancestor,
"Adam," lived only about 60,000 years ago. MtDNA and the Y
chromosome are independent parts of our genetic makeup and each tells a
different tale of successive genetic mutations over the eons. That is why
their approximate coalescence points are different. Yet while the dates
vary, both paths point emphatically to a surprisingly recent African origin
for all humans. The oldest known fossil remains of anatomically modern
humans were found in Ethiopia's
Omo River Valley.
The skeletons, known as Omo I and Omo II, have been dated to about 195,000
years ago. Although haplogroup L3 does not occur outside of Africa it is an important part of the human
migrations from that continent to the rest of the world. A single person of
the L3 lineage gave rise to the M and N haplogroups some 80,000 years ago.
All Eurasian mtDNA lineages are subsequently descended from these two
groups. The African ice age was characterized by drought rather than by
cold. But about 50,000 years ago a period of warmer temperatures and moist
climate made even parts of the arid Sahara
habitable. The climatic shift likely spurred hunter-gatherer migrations
into a steppe-like Sahara---and beyond.
This "Saharan Gateway" led humans out of Africa to the Middle East. The route they took is uncertain. They
may have traveled north down the Nile to
the Mediterranean coast and the Sinai. Alternatively, they may have crossed
what was then a land bridge connecting the Bab al Mandab to Arabia, after
which they either skirted the then-lush, verdant eastern coast of the Red
Sea or headed east along the Gulf of Aden towards the Arabian
Sea. When the climate again turned arid, expanding Saharan
sands slammed the Saharan Gateway shut. The desert was at its driest
between 20,000 and 40,000 years ago, and during this period MIddle East
immigrants became isolated from Africa.
From their new Middle East location,
however, they would go on to populate much of the world. N is a
macro-haplogroup descended from the African lineage L3. This line of
descent, with haplogroup M, traces the first human migrations out of Africa. The ancient members of N spawned sublineages
found across Eurasia and, eventually, the Americas. Early members of this
group lived in the eastern Mediterranean and Near East
region, where they likely coexisted for a time with pre-modern hominids
such as Neantertals. Excavations in Israel's Kebara cave (mount Carmel)
have unearthed Neandertal skeletons at least as recent as 60,000 years old.
Growing cognitive abilities likely gave these Upper Paleolithic humans
tremendous social advantages, evidenced by the appearance of modern thought
and behavior. This "great leap forward" may have enabled our
ancestors to outcompete and eventually replace evolutionary dead-end
lineages such as the Neandertals. The macro-haplogroup N is composed of
many subclades, which are often geographically distinct. Learning more about
these subclades will add further clarity to the big picture of human
genetic diversity, and is a primary goal of the Genographic Project.
Haplogroup R is descended from N and has since dispersed across much of the
globe. The lineage, in its many subgroups, appears on all continents except
Australia and Antarctica. Subgroups preHV, U, T, and J are found in
Europe and the Near East. The R5 and R6
lineages arose on the Indian subcontinent. Haplogroup K appeared some
16,000 years ago (on the R line of descent) when Europe's
glaciers finally began a retreat from their ice age maximum. Humans of the
era were living in the ice-free refuges of southern Europe--where
K is still found in its highest concentrations. As populations followed the
retreating ice northward, the lineage's descendents spread throughout most
of Europe. Tests have revealed that Otzi,
the 5200-year-old remains of a Copper Age man frozen in an Alpine glacier,
belongs to haplogroup K.
Analysis
of the mtDNA of Ötzi the Iceman, the frozen mummy from 3,300 BC found on
the Austrian-Italian border, has shown that Ötzi belongs to the K1
subcluster of the mitochondrial haplogroup K, but that it cannot be
categorized into any of the three modern branches of that subcluster.
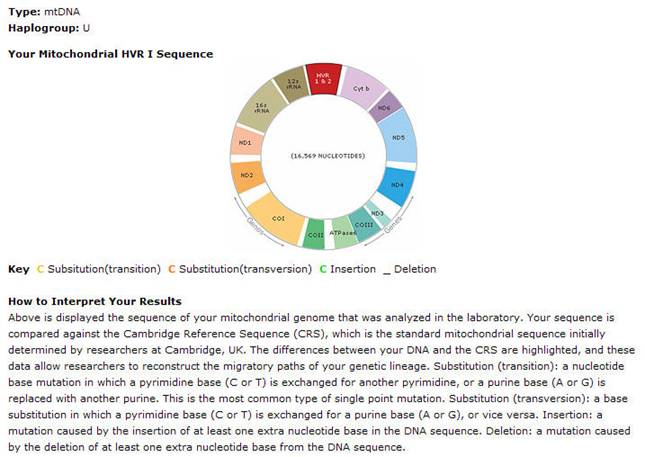
II. mtDNA
Sequence
|
mtDNA Haplogroup
|
U2b
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
HVR1 differences from CRS
|
16051G
|
16256T
|
16318G
|
|
|
|
|
|
|
|
|
|
|
|
|
HVR2 differences from CRS
|
73G
|
146C
|
263G
|
309.1C
|
309.2C
|
315.1C
|
|
|
|
|
|
|
|
|
|
Coding region
|
750G
|
1438G
|
1598A
|
1811G
|
3915A
|
4093G
|
|
|
4769G
|
5186T
|
7028T
|
8289G
|
8860G
|
11467G
|
|
|
11719A
|
11743T
|
11914A
|
12106T
|
12308G
|
12372A
|
|
|
13105G
|
13194A
|
14112A
|
14766T
|
15049T
|
15326G
|
The
mitochondrion sequenced in 1981 became known as the Cambridge Reference
Sequence (CRS) and has been used as a basis for comparison with your mtDNA.
In other words, any place in your mtDNA where you have a difference from
the CRS, is characterized as a mutation. if your results show no mutations
at all, it means that your mtDNA matches the CRS. A mutation happens a)
when a base replaces another base - for example a C (Cytosine) replaces an
A (Adenine), b) when a base is no longer in that position and c) when a new
base is inserted between the other bases without replacing any other. Those
mutations are represented below according to the following color code:
a)
Mutation at position: RED
b) Mutation Deleted: Strike
c) Mutation Inserted: Green
As
a way to make it possible to display all the positions that are included in
your test, please note that when you see "ATTCTAATTT" under
16010, it actually means that 16001 has an A (Adenine), 16002 has a T
(Thymine), 16003 has a T (Thymine), 16004 has a C (Cytosine) and so on...
Therefore, if your table of mutations above would show "192A" it
means that you should see the 3rd "C" under 16192 replaced by a
"A".
|
HVR1 Reference Sequence (Starts At: 16001)
|
|
16010
|
16020
|
16030
|
16040
|
16050
|
16060
|
16070
|
16080
|
|
ATTCTAATTT
|
AAACTATTCT
|
CTGTTCTTTC
|
ATGGGGAAGC
|
AGATTTGGGT
|
ACCACCCAAG
|
TATTGACTCA
|
CCCATCAACA
|
|
16090
|
16100
|
16110
|
16120
|
16130
|
16140
|
16150
|
16160
|
|
ACCGCTATGT
|
ATTTCGTACA
|
TTACTGCCAG
|
CCACCATGAA
|
TATTGTACGG
|
TACCATAAAT
|
ACTTGACCAC
|
CTGTAGTACA
|
|
16170
|
16180
|
16190
|
16200
|
16210
|
16220
|
16230
|
16240
|
|
TAAAAACCCA
|
ATCCACATCA
|
AAACCCCCTC
|
CCCATGCTTA
|
CAAGCAAGTA
|
CAGCAATCAA
|
CCCTCAACTA
|
TCACACATCA
|
|
16250
|
16260
|
16270
|
16280
|
16290
|
16300
|
16310
|
16320
|
|
ACTGCAACTC
|
CAAAGCCACC
|
CCTCACCCAC
|
TAGGATACCA
|
ACAAACCTAC
|
CCACCCTTAA
|
CAGTACATAG
|
TACATAAAGC
|
|
16330
|
16340
|
16350
|
16360
|
16370
|
16380
|
16390
|
16400
|
|
CATTTACCGT
|
ACATAGCACA
|
TTACAGTCAA
|
ATCCCTTCTC
|
GTCCCCATGG
|
ATGACCCCCC
|
TCAGATAGGG
|
GTCCCTTGAC
|
|
16410
|
16420
|
16430
|
16440
|
16450
|
16460
|
16470
|
16480
|
|
CACCATCCTC
|
CGTGAAATCA
|
ATATCCCGCA
|
CAAGAGTGCT
|
ACTCTCCTCG
|
CTCCGGGCCC
|
ATAACACTTG
|
GGGGTAGCTA
|
|
16490
|
16500
|
16510
|
16520
|
16530
|
16540
|
16550
|
16560
|
|
AAGTGAACTG
|
TATCCGACAT
|
CTGGTTCCTA
|
CTTCAGGGTC
|
ATAAAGCCTA
|
AATAGCCCAC
|
ACGTTCCCCT
|
TAAATAAGAC
|
|
16569
|
|
|
|
|
|
|
|
|
ATCACGATG
|
|
|
|
|
|
|
|
|
HVR2 Reference Sequence (Starts
At: 1)
|
|
10
|
20
|
30
|
40
|
50
|
60
|
70
|
80
|
|
GATCACAGGT
|
CTATCACCCT
|
ATTAACCACT
|
CACGGGAGCT
|
CTCCATGCAT
|
TTGGTATTTT
|
CGTCTGGGGG
|
GTATGCACGC
|
|
90
|
100
|
110
|
120
|
130
|
140
|
150
|
160
|
|
GATAGCATTG
|
CGAGACGCTG
|
GAGCCGGAGC
|
ACCCTATGTC
|
GCAGTATCTG
|
TCTTTGATTC
|
CTGCCTCATC
|
CTATTATTTA
|
|
170
|
180
|
190
|
200
|
210
|
220
|
230
|
240
|
|
TCGCACCTAC
|
GTTCAATATT
|
ACAGGCGAAC
|
ATACTTACTA
|
AAGTGTGTTA
|
ATTAATTAAT
|
GCTTGTAGGA
|
CATAATAATA
|
|
250
|
260
|
270
|
280
|
290
|
300
|
310
|
320
|
|
ACAATTGAAT
|
GTCTGCACAG
|
CCACTTTCCA
|
CACAGACATC
|
ATAACAAAAA
|
ATTTCCACCA
|
AACCCCCCCCCT
|
CCCCCCGCTTC
|
|
330
|
340
|
350
|
360
|
370
|
380
|
390
|
400
|
|
TGGCCACAGC
|
ACTTAAACAC
|
ATCTCTGCCA
|
AACCCCAAAA
|
ACAAAGAACC
|
CTAACACCAG
|
CCTAACCAGA
|
TTTCAAATTT
|
|
410
|
420
|
430
|
440
|
450
|
460
|
470
|
480
|
|
TATCTTTTGG
|
CGGTATGCAC
|
TTTTAACAGT
|
CACCCCCCAA
|
CTAACACATT
|
ATTTTCCCCT
|
CCCACTCCCA
|
TACTACTAAT
|
|
490
|
500
|
510
|
520
|
530
|
540
|
550
|
560
|
|
CTCATCAATA
|
CAACCCCCGC
|
CCATCCTACC
|
CAGCACACAC
|
ACACCGCTGC
|
TAACCCCATA
|
CCCCGAACCA
|
ACCAAACCCC
|
|
570
|
580
|
|
|
|
|
|
|
|
AAAGACACCC
|
CCCA
|
|
|
|
|
|
|
III. mtDNA
Haplogroup Maps
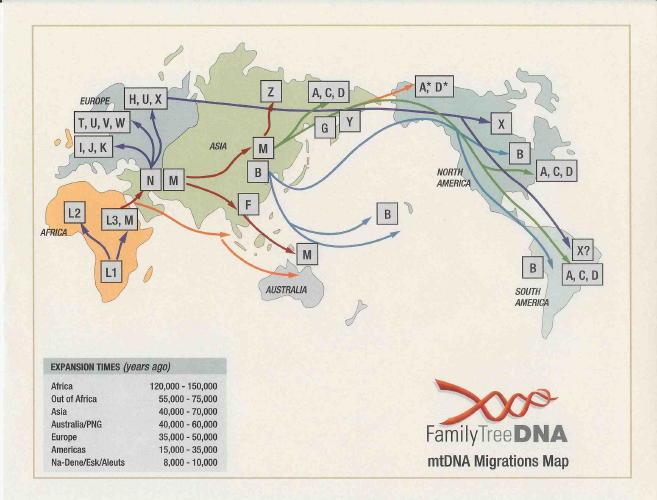
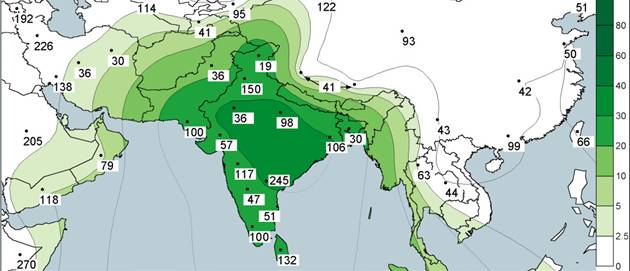
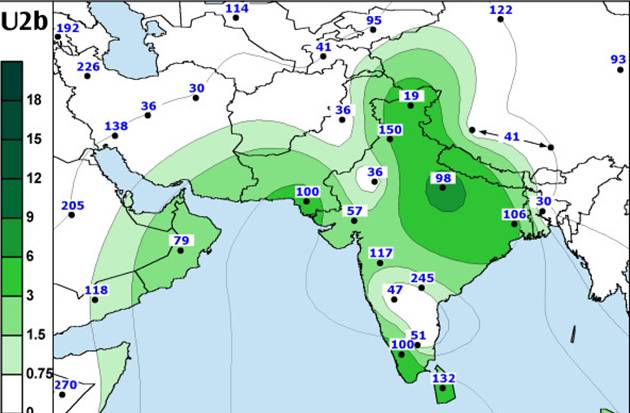
Subhaplogroup
U2 distribution (above); subgroup haplogroup U2b distribution (below)
From
National Geographic:
Your
Branch on the Human Family Tree
Your
DNA results identify you as belonging to a specific branch of the human
family tree called haplogroup U. Haplogroup U contains the following subgroups:
U*, U1, U1a, U1b, U2, U3, U4, U7.
The
map above shows the direction that your maternal ancestors took as they set
out from their original homeland in East Africa.
While humans did travel many different paths during a journey that took
tens of thousands of years, the lines above represent the dominant trends
in this migration.
Over
time, the descendants of your ancestors ultimately made it into
northeastern Europe, where most members of
your haplogroup are found today. But before we can take you back in time
and tell their stories, we must first understand how modern science makes
this analysis possible.
How
DNA Can Help
(To
follow along, click See Your DNA Analysis above to view the data produced
from your cheek scraping.)
The string
of 569 letters shown above is your mitochondrial sequence, with the letters
A, C, T, and G representing the four nucleotides—the chemical building
blocks of life—that make up your DNA. The numbers at the top of the page
refer to the positions in your sequence where informative mutations have
occurred in your ancestors, and tell us a great deal about the history of
your genetic lineage.
Here's
how it works. Every once in a while a mutation—a random, natural (and
usually harmless) change—occurs in the sequence of your mitochondrial DNA.
Think of it as a spelling mistake: one of the "letters" in your
sequence may change from a C to a T, or from an A to a G.
(Explore
the Genetics Overview to learn more about population genetics.)
After
one of these mutations occurs in a particular woman, she then passes it on
to her daughters, and her daughters' daughters, and so on. (Mothers also
pass on their mitochondrial DNA to their sons, but the sons in turn do not
pass it on.)
Geneticists
use these markers from people all over the world to construct one giant
mitochondrial family tree. As you can imagine, the tree is very complex,
but scientists can now determine both the age and geographic spread of each
branch to reconstruct the prehistoric movements of our ancestors.
By
looking at the mutations that you carry, we can trace your lineage,
ancestor by ancestor, to reveal the path they traveled as they moved out of
Africa. Our story begins with your
earliest ancestor. Who was she, where did she live, and what is her story?
(Click
Explore Your Route Map on the right side of the page to return to the map
showing your haplogroup's ancestral journey.)
Your
Ancestral Journey: What We Know Now
We
will now take you back through the stories of your distant ancestors and
show how the movements of their descendants gave rise to your mitochondrial
lineage.
Each
segment on the map above represents the migratory path of successive groups
that eventually coalesced to form your branch of the tree. We start with
your oldest ancestor, "Eve," and walk forward to more recent
times, showing at each step the line of your ancestors who lived up to that
point.
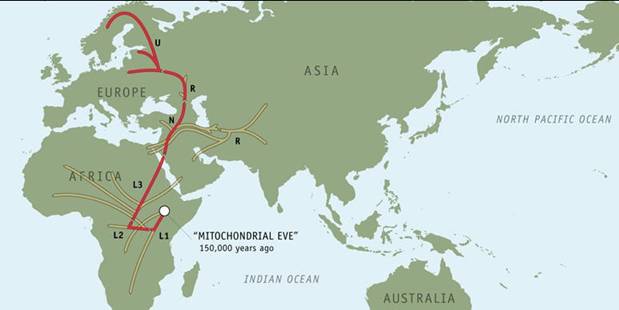
Mitochondrial
Eve: The Mother of Us All
Ancestral
Line: "Mitochondrial Eve"
Our
story begins in Africa sometime between
150,000 and 170,000 years ago, with a woman whom anthropologists have
nicknamed "Mitochondrial Eve."
She
was awarded this mythic epithet in 1987 when population geneticists
discovered that all people alive on the planet today can trace their maternal
lineage back to her.
But
Mitochondrial Eve was not the first female human. Homo sapiens evolved in Africa around 200,000 years ago, and the first
hominids—characterized by their unique bipedal stature—appeared nearly two
million years before that. Yet despite humans having been around for almost
30,000 years, Eve is exceptional because hers is the only lineage from that
distant time to survive to the present day.
Which
begs the question, "So why Eve?"
Simply
put, Eve was a survivor. A maternal line can become extinct for a number of
reasons. A woman may not have children, or she may bear only sons (who do
not pass her mtDNA to the next generation). She may fall victim to a
catastrophic event such as a volcanic eruption, flood, or famine, all of which
have plagued humans since the dawn of our species.
None
of these extinction events happened to Eve's line. It may have been simple
luck, or it may have been something much more. It was around this same time
that modern humans' intellectual capacity underwent what author Jared
Diamond coined the Great Leap Forward. Many anthropologists believe that
the emergence of language gave us a huge advantage over other early human
species. Improved tools and weapons, the ability to plan ahead and
cooperate with one another, and an increased capacity to exploit resources
in ways we hadn't been able to earlier, all allowed modern humans to
rapidly migrate to new territories, exploit new resources, and outcompete
and replace other hominids, such as the Neandertals.
It
is difficult to pinpoint the chain of events that led to Eve's unique
success, but we can say with certainty that all of us trace our maternal
lineage back to this one woman.
The
L Haplogroups: The Deepest Branches
Ancestral
line: "Eve" > L1/L0
Mitochondrial
Eve represents the root of the human family tree. Her descendents, moving
around within Africa, eventually split
into two distinct groups, characterized by a different set of mutations
their members carry.
These
groups are referred to as L0 and L1, and these individuals have the most
divergent genetic sequences of anybody alive today, meaning they represent
the deepest branches of the mitochondrial tree. Importantly, current
genetic data indicates that indigenous people belonging to these groups are
found exclusively in Africa. This means
that, because all humans have a common female ancestor, "Eve,"
and because the genetic data shows that Africans are the oldest groups on
the planet, we know our species originated there.
Haplogroups
L1 and L0 likely originated in East Africa
and then spread throughout the rest of the continent. Today, these lineages
are found at highest frequencies in Africa's
indigenous populations, the hunter-gatherer groups who have maintained
their ancestors' culture, language, and customs for thousands of years.
At
some point, after these two groups had coexisted in Africa
for a few thousand years, something important happened. The mitochondrial
sequence of a woman in one of these groups, L1, mutated. A letter in her
DNA changed, and because many of her descendants have survived to the
present, this change has become a window into the past. The descendants of
this woman, characterized by this signpost mutation, went on to form their
own group, called L2. Because the ancestor of L2 was herself a member of
L1, we can say something about the emergence of these important groups: Eve
begat L1, and L1 begat L2. Now we're starting to move down your ancestral
line.
Haplogroup
L2: West Africa
Ancestral
line: "Eve" > L1/L0 > L2
L2 individuals
are found in sub-Saharan Africa, and like their L1 predecessors, they also
live in Central Africa and as far south as South Africa. But whereas L1/L0
individuals remained predominantly in eastern and southern Africa, your ancestors broke off into a different
direction, which you can follow on the map above.
L2
individuals are most predominant in West Africa,
where they constitute the majority of female lineages. And because L2
individuals are found at high frequencies and widely distributed along
western Africa, they represent one of the
predominant lineages in African-Americans. Unfortunately, it is difficult
to pinpoint where a specific L2 lineage might have arisen. For an
African-American who is L2—the likely result of West Africans being brought
to America during the
slave trade—it is difficult to say with certainty exactly where in Africa that lineage arose.
Fortunately,
collaborative sampling with indigenous groups is currently underway to help
learn more about these types of questions and to possibly bridge the gap
that was created during those transatlantic voyages hundreds of years ago.
Haplogroup
L3: Out of Africa
Ancestral
line: "Eve" > L1/L0 > L2 > L3
Your
next signpost ancestor is the woman whose birth around 80,000 years ago
began haplogroup L3. It is a similar story: an individual in L2 underwent a
mutation to her mitochondrial DNA, which was passed onto her children. The
children were successful, and their descendants ultimately broke away from
the L2 clan, eventually separating into a new group called L3. You can see
above that this has revealed another step in your ancestral line.
While
L3 individuals are found all over Africa,
including the southern reaches of sub-Sahara, L3 is important for its
movements north. You can follow this movement of the map above, seeing
first the expansions of L1/L0, then L2, and followed by the northward
migration of L3.
Your
L3 ancestors were significant because they are the first modern humans to
have left Africa, representing the deepest
branches of the tree found outside of that continent.
Why
would humans have first ventured out of the familiar African hunting
grounds and into unexplored lands? It is likely that a fluctuation in
climate may have provided the impetus for your ancestors' exodus out of Africa.
The
African Ice Age was characterized by drought rather than by cold. Around
50,000 years ago the ice sheets of northern Europe began to melt,
introducing a period of warmer temperatures and moister climate in Africa. Parts of the inhospitable Sahara
briefly became habitable. As the drought-ridden desert changed to savanna,
the animals your ancestors hunted expanded their range and began moving
through the newly emerging green corridor of grasslands. Your nomadic
ancestors followed the good weather and plentiful game northward across
this Saharan Gateway, although the exact route they followed remains to be
determined.
Today,
L3 individuals are found at high frequencies in populations across North Africa. From there, members of this group went
in a few different directions. Some lineages within L3 testify to a
distinct expansion event in the mid-Holocene that headed south, and are
predominant in many Bantu groups found all over Africa.
One group of individuals headed west and is primarily restricted to
Atlantic western Africa, including the
islands of Cabo Verde.
Other
L3 individuals, your ancestors, kept moving northward, eventually leaving
the African continent completely. These people currently make up around ten
percent of the Middle Eastern population, and gave rise to two important
haplogroups that went on to populate the rest of the world.
Haplogroup
N: The Incubation Period
Ancestral
line: "Eve" > L1/L0 > L2 > L3 > N
Your
next signpost ancestor is the woman whose descendants formed haplogroup N.
Haplogroup N comprises one of two groups that were created by the
descendants of L3.
The
first of these groups, M, was the result of the first great wave of
migration of modern humans to leave Africa.
These people likely left the continent across the Horn of Africa near Ethiopia, and their descendants followed a
coastal route eastward, eventually making it all the way to Australia and Polynesia.
The
second great wave, also of L3 individuals, moved north rather than east and
left the African continent across the Sinai Peninsula, in present-day Egypt. Also
faced with the harsh desert conditions of the Sahara, these people likely
followed the Nile basin, which would have
proved a reliable water and food supply in spite of the surrounding desert
and its frequent sandstorms.
Descendants
of these migrants eventually formed haplogroup N. Early members of this
group lived in the eastern Mediterranean region and western Asia, where they likely coexisted for a time with
other hominids such as Neandertals. Excavations in Israel's Kebara
Cave (Mount
Carmel) have unearthed Neandertal skeletons as recent as
60,000 years old, indicating that there was both geographic and temporal
overlap of these two hominids.
The
ancient members of haplogroup N spawned many sublineages, which spread
across much of the rest of the globe and are found throughout Asia, Europe,
India, and the Americas.
Haplogroup
R: Spreading Out
Ancestral
line: "Eve" > L1/L0 > L2 > L3 > N > R
After
several thousand years in the Near East,
individuals belonging to a new group called haplogroup R began to move out
and explore the surrounding areas. Some moved south, migrating back into
northern Africa. Others went west across
Anatolia (present-day Turkey)
and north across the Caucasus Mountains of Georgia and southern Russia.
Still others headed east into the Middle East, and on to Central
Asia. All of these individuals had one thing in common: they
shared a female ancestor from the N clan, a recent descendant of the
migration out of Africa.
The
story of haplogroup R is complicated, however, because these individuals
can be found almost everywhere, and because their origin is quite ancient.
In fact, the ancestor of haplogroup R lived relatively soon after humans
moved out of Africa during the second
wave, and her descendants undertook many of the same migrations as her own
group, N.
Because
the two groups lived side by side for thousands of years, it is likely that
the migrations radiating out from the Near East
comprised individuals from both of these groups. They simply moved
together, bringing their N and R lineages to the same places around the
same times. The tapestry of genetic lines became quickly entangled, and
geneticists are currently working to unravel the different stories of
haplogroups N and R, since they are found in many of the same far-reaching
places.
Haplogroup
U: Your Branch on the Tree
Ancestral
line: "Eve" > L1/L0 > L2 > L3 > N > R > U
We
finally arrive at your own clan, a group of individuals who descend from a
woman in the R branch of the tree. Because of the great genetic diversity
found in haplogroup U, it is likely that she lived around 50,000 years ago.
Her
descendants gave rise to several different subgroups, some of which exhibit
very specific geographic homelands. The very old age of these subgroups has
led to a wide distribution; today they harbor specific European, northern
African, and Indian components, and are found in Arabia, the northern
Caucasus Mountains, and throughout the Near East.
One
of these subgroups, U5, is restricted to Finland in its variation. This
is likely the result of the significant geographical, linguistic, and cultural
isolation of the Finnish populations, which would have restricted
geographic distribution of this subgroup and kept it fairly isolated
genetically. The Saami, reindeer hunters who follow the herds from Siberia
to Scandinavia each season, also have U5 lineages in their population,
indicating that these may have been introduced during their movements into
these northern territories.
The U5 lineage is found as far south as the Near East,
though at much smaller frequencies and at much lower diversity. Because
these individuals contain lineages that first evolved in Europe, their
presence in the Near East is the result of a back-migration of people who
left northern Europe and headed south, as
though retracing the migratory paths of their own ancestors.
Another
interesting subgroup is U6, which branched off from haplogroup R while
still in the Middle East. This subgroup
moved southward and today is found in parts of northern Africa.
Today U6 individuals are found in around ten percent of people living in North Africa. Despite the great geographic distances
between subgroups U5 and U6, all members still share the same maternal
ancestral line that gave rise to your ancestral clan, haplogroup U.
While
some members of your haplogroup headed north into Scandinavia, or south
into North Africa, most members of your haplogroup U stem from a group that
moved northward out of the Near East. These women crossed the rugged
Caucasus Mountains in southern Russia,
and moved on to the steppes of the Black Sea.
These individuals represent movements from the Black Sea steppes west into
regions that comprise the present-day Baltic States and western Eurasia. This grassland then served as the home base
for subsequent movements north and west. Today, members of these lineages are
found in Europe and the eastern Mediterranean
at frequencies of almost seven percent of the population.
Anthropology
vs. Genealogy
DNA
markers require a long time to become informative. While mutations occur in
every generation, it requires at least hundreds—normally thousands—of years
for these markers to become windows back into the past, signposts on the
human tree.
Still,
our own genetic sequences often reveal that we fall within a particular
sub-branch, a smaller, more recent branch on the tree.
While
it may be difficult to say anything about the history of these sub-groups,
they do reveal other people who are more closely related to us. It is a
useful way to help bridge the anthropology of population genetics with the
genealogy to which we are all accustomed.
One
of the ways you can bridge this gap is to compare your own genetic lineage
to those of people living all over the world. Mitosearch.org is a database
that allows you to compare both your genetic sequence as well as your
surname to those of thousands of people who have already joined the
database. This type of search is a valuable way of inferring population
events that have occurred in more recent times (i.e., the past few hundred
years).
Looking
Forward (Into the Past): Where Do We Go From Here?
Although
the arrow of your haplogroup currently ends in Northern Europe and
Scandinavia, this isn't the end of the journey for haplogroup U.
This is where the genetic clues get murky and your DNA trail goes cold.
Your initial results shown here are based upon the best information
available today—but this is just the beginning.
A
fundamental goal of the Genographic Project is to extend these arrows
further toward the present day. To do this, Genographic has brought
together ten renowned scientists and their teams from all over the world to
study questions vital to our understanding of human history. By working
together with indigenous peoples around the globe, we are learning more
about these ancient migrations.
Y-DNA
|